In the early stages of a pandemic like Covid-19, public health officials need a lot of answers fast. How quickly is the virus spreading, and through which routes? How can we contain it? And when can we safely relax the most stringent control measures such as shelter-in-place?
Answering those questions is never easy, but in the face of the new coronavirus, epidemiologists have a powerful tool that wasn’t available for the earlier SARS and MERS epidemics (also caused by coronaviruses): rapid, large-scale sequencing of viral genomes. These genetic sequences from viruses that have infected patients, together with old-fashioned tracing of personal contacts, allow health officials to track the spread of a virus from person to person and place to place faster and more accurately than ever before. That speed, they hope, will translate into earlier control of the virus, and more precise management of the pandemic’s end stages.
Geneticists have been able to sequence viral genomes for decades, of course — but the latest advances in the technology mean they can now do so in a matter of hours or days. Just as quickly, scientists around the world can share what they learn via a global open-source network known as Nextstrain. That speed and cooperation have been a game-changer, enabling this “genomic epidemiology” to be used in real time as the Covid-19 pandemic unfolds.
“We have used genomic epidemiology in other contexts where we were getting sequence in a month or a few weeks, but we’ve never had anything where we’ve had such fast turnaround or the number of sequences being shared from so many places so quickly,” says Emma Hodcroft, a genetic epidemiologist at the University of Basel in Switzerland and member of the Nextstrain network.
Using genome sequences, researchers can deduce evolutionary relationships between different versions of the virus, helping to track the origin of a pandemic. From this and other information, they can reconstruct how and where the virus may have spread from person to person.
Sloppy copies
Much of the power of genomic epidemiology stems from the fact that most viruses make lots of mistakes when they copy their genomes, so changes in the sequence — that is, new mutations — turn up relatively often. That’s especially true of viruses that use RNA as their genetic material, as coronaviruses do. Very few of these mutations affect how the virus behaves — most have no apparent consequence at all — but researchers can use them as markers to build a family tree of the virus and to see how the virus has changed over time and how it has spread from locale to locale.
Early in the Covid-19 outbreak, researchers all over the world began sequencing viruses sampled from patients and building a family tree of the virus on Nextstrain. Almost immediately, they could see that the tree was short — the virus sequences had not yet accumulated many distinct mutations, meaning that the new coronavirus, SARS-CoV-2, hadn’t been infecting humans for long. Moreover, the tree had a single trunk, indicating that every virus infecting humans likely descended from a single case in early December 2019.
In contrast, periodic outbreaks of MERS in humans in the 2010s look more like a shrubland: multiple small clusters of virus genotypes that are more closely related to camel viruses than to one another, indicating that MERS must have jumped repeatedly from camels to humans and then fizzled out.
The SARS-CoV-2 virus’s genetic mutability also means that epidemiologists can use changes in its genome to trace the spread of the virus during an epidemic. That’s because most mutations are essentially random, so each branch of the virus tree is likely to bear its own unique set of mutations. If one person’s virus contains mutations A, B and C, for example, that person could have caught it from someone whose virus carries A and B or A and C, but not from someone whose virus has A, B, C and D.
Mutations in a viral genome can serve as genetic breadcrumbs, giving scientists insight into viral origins and spread.
Early in the current pandemic, Nextstrain noted the appearance of identical or near-identical coronavirus genomes from people in countries as widely spaced as Canada, Australia and the UK. The genomes were so similar that scientists inferred they must have shared a common source. That red flag prompted further questioning, which revealed that all of the sick had recently travelled to Iran.
“We could confirm that these patients must have been infected in Iran, because that’s the only thing they had in common,” says Hodcroft. Without the genomes, nothing would have linked those patients, and the Iranian connection would not have been noticed as quickly. Similarly, most viral genomes in the New York City region closely match those seen earlier in Europe, suggesting that infections came from there, not directly from China.
Of course, epidemiologists also track transmission routes the traditional way, by interviewing people and tracing their contacts. However, this method can’t keep up in the face of a pandemic, where thousands of new cases are added every day.
“There’s an advantage to old-fashioned shoe-leather contact tracing, because you can actually talk to people and find out who they spoke to,” says Hodcroft. “But as the number of cases rises, you cannot contact-trace everyone. You just don’t have enough people. That’s where using genetics can be a big help.”
Viral family tree
Genomes can be especially good at answering a key public health question early in an epidemic: Are new infections in a given locality imported by travelers, or are they homegrown? The latter — the result of the virus circulating within the community — would create a need for the social-distancing measures now familiar to so many of us.
“If you’re seeing strains that are really, really similar, that suggests that they’re transmitting locally,” says Shirlee Wohl, a genomic epidemiologist at Johns Hopkins Bloomberg School of Public Health and coauthor of a review of the field in the 2016 Annual Review of Virology. “That’s information you really can’t get from any other method.”
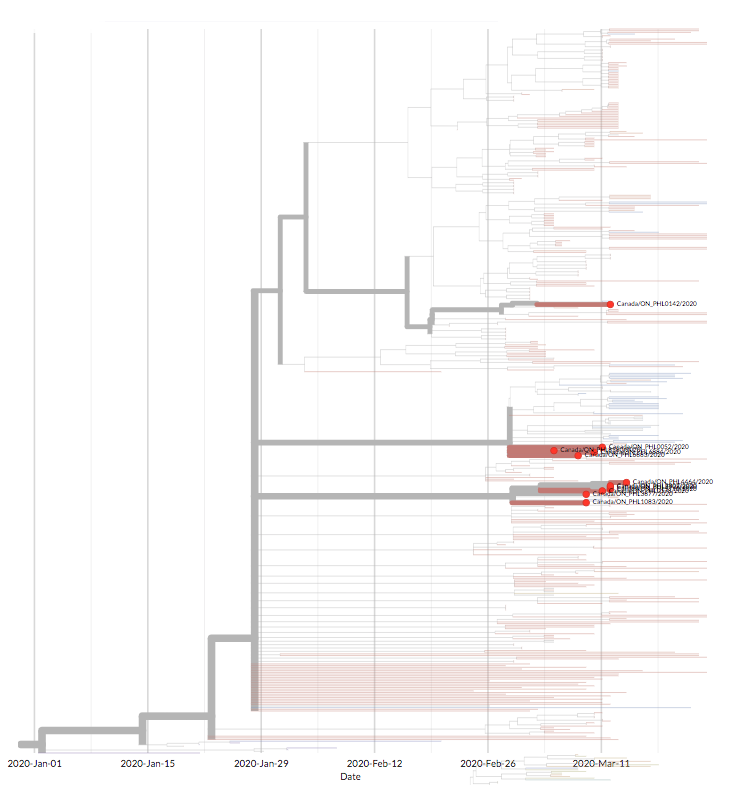
This portion of the evolutionary tree of SARS-CoV-2 virus shows three separate clusters of virus from Covid-19 patients in Ontario, Canada (red dots). Within each cluster, viruses are closely related, indicating local transmission, but the three clusters are more distantly related, indicating that each cluster was introduced separately from elsewhere. The most likely source is the US, based on the similarities in the viral sequences.
CREDIT: NEXTSTRAIN.ORG
For example, the first Covid-19 infection in the state of Washington was in a traveler returning from Wuhan, China, where the outbreak began. When a later infection in Washington turned out to have a nearly identical sequence, this was strong evidence of community transmission — especially because the two individuals, though unacquainted, lived in the same county.
Unfortunately for genetic detectives, the Covid-19 virus changes a little too slowly for optimal tracking of transmission chains, Wohl notes. HIV, in contrast, mutates so quickly that each person usually carries a unique genotype, allowing epidemiologists to pinpoint the exact source of each new infection. For the Covid-19 virus, each viral lineage accumulates about 30 new mutations per year, which works out to about one new mutation per two links in the transmission chain. As a result, exactly the same viral genome sequence can be found in several people, so genome-trackers can narrow transmission down only to a handful of suspects.
Additional uncertainty comes from the fact that researchers can’t possibly sequence viruses from every infected individual in a widespread pandemic. As of April 20, nearly 2.5 million people worldwide had been infected with SARS-CoV-2, but Nextstrain listed just 4,558 sequences. That can lead to false conclusions. “The beautiful danger is it looks like it can tell you a lot of enticing stories,” says Hodcroft. “But we don’t know that the scenario is exactly what happened.”
In late February, for example, sequencers found patients in Germany and Italy who shared the same unusual viral mutation. Since the German patient had gotten sick sooner, this led some researchers to suggest that the virus had spread from Germany to Italy. In reality, though, both German and Italian patients could have caught the virus from some third person, yet unidentified, whose virus was not sequenced.
Still, these limitations have not kept genomic epidemiology from playing a key role in the Covid-19 pandemic. The approach has helped public health officials identify the pathogen, trace its travels and recognize community spread promptly. And in the months ahead, the method may have more to contribute.
Using virus sequence data, researchers can track the spread of Covid-19 around the world. The animation starts in late 2019 and shows the first virus genome sequences found in January 2020 from Wuhan, China, with disease spreading rapidly in the weeks after.
CREDIT: NEXTSTRAIN.ORG
One contribution is likely to come from longer-term studies of where mutations fall in the genome. Most of the genetic changes, remember, make little or no difference to the virus: They are “neutral,” in evolutionary biologists’ parlance. But mutations that change the shape of key proteins, such as the spike protein on the surface of the virus that binds to receptors in our cells, are more likely to matter.
Looking to see how these regions have changed since the virus infected humans may eventually help virologists understand why this particular virus has been able to adapt to us so well, says Hodcroft. However, this will require painstaking experiments over many months to reveal the functional effect of each mutation. “It’s not something that’s done in an afternoon,” she says.
Before that happens, genomic epidemiology promises to help public health officials find the smartest way to relax the burdensome social-distancing measures that are so important in controlling the pandemic right now. By using genomic breadcrumbs to track the transmission of the virus, epidemiologists hope to identify which activities are most likely to spread the virus. If schools, for example, turn out to pose a relatively low risk, authorities may be able to re-open those sooner.
“That hopefully means we can start relaxing those lockdowns faster than we might have 10 years ago, when we didn’t have this technology,” says Hodcroft. But that depends on a key factor that was not much in evidence at the start of the epidemic: the willingness of politicians to heed scientists’ warnings and advice.