There are roughly 37 trillion cells in the human body, and each keeps us alive in some small way. There are cells that ward off disease, cells that absorb nutrients from our lunch, and cells that keep our hearts pumping and brains humming.
No two cells in a body are exactly alike — even though they’re by and large genetically identical, and even when we’re talking about two cells of the same type. They differ in precisely which genes within them are active and how efficiently those genetic instructions are used to make RNA and protein.
Yet in studying cells, scientists generally take thousands to millions of cells and mash them up. That’s a great way to study average cell behavior. But it stands to miss a lot.
Today, new technologies allow scientists to capture the nuance that’s lost in bulk populations. Using single-cell DNA sequencing, studying gene activation in individual cells or simply peering through the microscope, they are making startling new observations about what cells are doing.
These techniques have the potential to “answer deep biological questions related to development, aging, the relationship between health and disease — all super-important areas,” says Stephen Quake, a Stanford biophysicist who coauthored a 2011 article on single-cell genomics in the Annual Review of Genetics. Here are three intriguing findings, with the promise of many more to come.
Finding new cell types
This may come as a surprise: We still don’t know all the types of cells in the human body. That’s because many molecular techniques have been ill-suited for uncovering rare cell types, says Aviv Regev, a computational and systems biologist at the Broad Institute of MIT and Harvard.
Regev likens studying large populations of cells to making a fruit smoothie with plenty of bananas and strawberries but just a few blueberries. Take one sip and you’ll taste the bananas and strawberries — but you’ll probably never pick out the blueberries, no matter how discerning your palate. Finding rare cell types presents a similar challenge.
“The only way to get them would be if you search for them specifically,” Regev says. “But how could you do so if you did not already know they exist?”
That’s where single-cell analysis comes in. In one such study, Regev and a team of researchers at Harvard and Massachusetts General Hospital analyzed nearly 7,500 individual cells from the airways of mice with a technique known as single-cell RNA-seq. The method involves sequencing the messenger RNA molecules contained in a cell. Messenger RNA relays the genetic instructions in DNA to the place in cells where proteins are made, so the technique lets scientists see just which genes are active in a cell and how active they are.
“We find those rare ‘blueberries’ and study their properties without them getting lost in the mix,” Regev says.
Using powerful algorithms to analyze the single-cell RNA-seq data, Regev and her colleague Jay Rajagopal led one of two independent teams to discover a new, rare lung cell type that they named a pulmonary ionocyte. Importantly, they noticed a highly active gene in these pulmonary ionocytes — one that codes for a protein that malfunctions in cystic fibrosis.
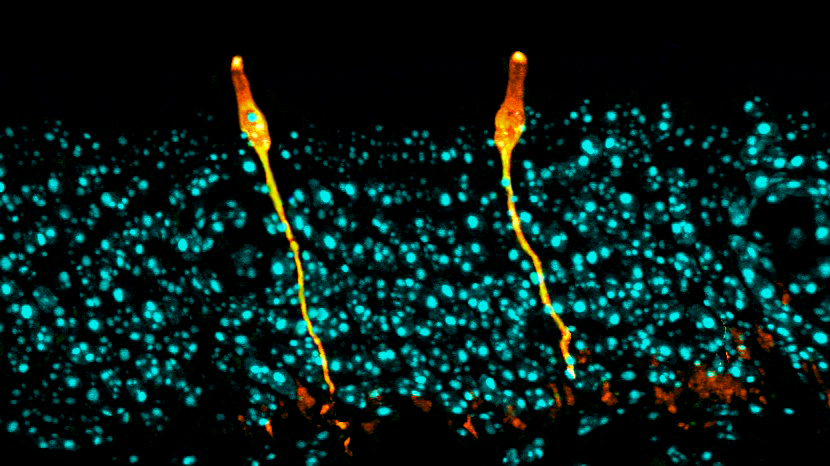
Two independent teams of researchers used single-cell analysis to discover a new cell type: the pulmonary ionocyte. These cells, shown here in orange in the respiratory tract of a mouse, produce large quantities of a receptor that malfunctions in people with cystic fibrosis.
CREDIT: D.T. MONTORO ET AL / NATURE 2018
The protein sits in the cell membrane and allows chloride ions to flow outward. But in patients with the disease, the protein is either defective, not produced or made in smaller amounts, causing thick and sticky mucus to build up in the airways and creating a perfect bacterial breeding ground. Drugs can treat certain mutant protein versions; knowing what cells make the protein might allow scientists to one day deliver treatments more precisely.
Regev has since collaborated with other researchers and found new subtypes of cells in the nervous system and gut, as well as cell types that show up during diseases in places they usually don’t. These, too, could offer leads to more targeted therapies, she says.
Nevertheless, they persisted
When a cell divides, it creates two genetically identical daughter cells — but that doesn’t guarantee that those cells will behave in the same way. Army doctor Joseph Bigger noticed a curious example of this happening in the 1940s after treating bacteria with antibiotics.
Bigger observed that most of the bacteria died, but sometimes a few cells hung around. The most obvious reason was that the survivors were genetically resistant to the drug, but Bigger found that most descendants of these bacteria were drug-sensitive. If the surviving bacteria had carried an antibiotic-resistance gene, all of the descendant cells would have been drug-resistant, so something else was going on.
This phenomenon is known as persistence, and researchers have since shown that these persister bacteria are slow-growing, which allows them to withstand drugs that target cells with an active metabolism.
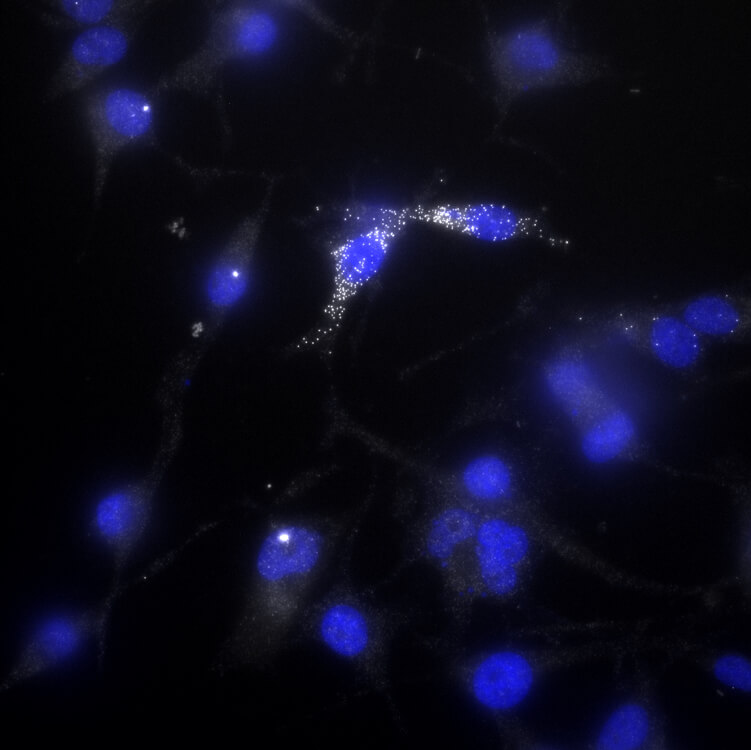
Melanoma cells that activate AXL, a drug-resistance gene, are more likely to survive exposure to the anti-melanoma drug vemurafenib. One such “persister” cell is shown in this image — the white dots indicate AXL activation.
CREDIT: BEN EMERT
Persistence isn’t genetically hardwired into the cells, but arises from natural variation in gene activation from one cell to the next. And that is helpful for bacteria. If all of them were slow-growing persisters, the population would fall behind on its main objective: to be fruitful and multiply. But without any persisters, the bacteria risk being wiped out by an antibiotic assault. The bet-hedging of persistence allows bacteria to survive in ever-changing environments.
Some cancer cells use a similar, though distinct, strategy. Sydney Shaffer, a cancer biologist at the University of Pennsylvania, drew inspiration from bacterial persistence studies to understand why certain tumor cells survive drug treatment while others don’t. “It’s fundamentally a problem that occurs at the single-cell level,” she says.
Shaffer found that melanoma cells can survive an anti-melanoma drug called vemurafenib without acquiring a permanent genetic change. To understand what was going on, she used fluorescent molecular probes to see whether cells that survive the drug had already activated known drug-resistance genes prior to exposure — which turned out to be true.
The whole dish would be dark, she says, “until you found one of these persister cells — and it would be lit up.”
Shaffer and her colleagues have found that persister cells pre-activate multiple drug-resistance genes, which suggests that the activity of these genes is not random but somehow coordinated. Understanding what signals control tumor persistence could lead to more effective combination therapies, with one drug to kill the majority of tumor cells and a second to root out persisters.
Out of one, many
Each cell in the body has the same genetic code, yet these cells take on different roles. The neurons that fire in your brain look and act nothing like the cells that process toxins in your liver, for example.
Scientists have spent decades identifying genes that regulate this dizzying diversification of cells. But Allon Klein, a systems biologist at Harvard, is taking a genome-wide approach to speed up discovery. Klein and collaborators developed a method that streams single cells assembly-line-style and measures their RNA. This allows scientists to quickly look at all the genes active in a cell rather than homing in on a favorite few — in thousands of cells at a time.
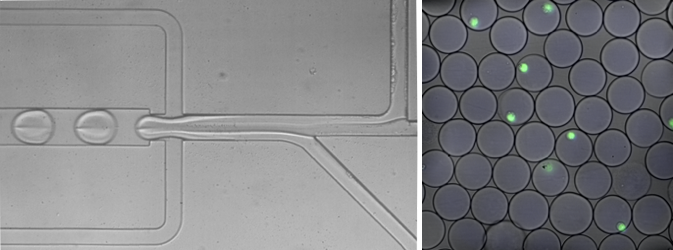
Harvard researchers have developed a fast-flowing, assembly-line-style approach to measure the genes that are active in each cell. On the left, cells moving through a narrow tube merge with liquid droplets that contain the reagents needed to measure the cells’ gene activity. On the right, cells individually encased in droplets are shown in green.
CREDIT: IGNAS MAZELIS
“We have essentially opened the floodgates,” Klein says. “You no longer need a five-year project to find one difference. You just run it in a day. So it’s really been transformational.”
In 2018, Klein’s team used the method to study zebrafish — small tropical fish often used in developmental studies. They isolated single cells from zebrafish embryos during their first 24 hours to map out how those early cells go on to form different tissues.
Open a biology textbook and you’ll see development depicted as a roadmap. A cell reaches a fork in the road and can take several possible paths to arrive at different final states. But Klein’s team found that this was too simplistic: In reality, a cell’s lineage doesn’t necessarily determine its destiny. One example, the scientists found, was the pharyngeal arches, a series of embryonic tissues that form the face, jaw and other parts of the head and neck. The team, reporting in Science, found that these arches contained cells from multiple lineages that had converged on a common fate — an outcome that current, simpler models can’t explain.
Science pointed to similar work Klein did as an example of why it declared tracking development at the single-cell level its 2018 Breakthrough of the Year. Researchers are already using the single-cell genome-wide approach to characterize beta cells, which produce insulin in the pancreas but are destroyed by the immune system in type 1 diabetes. Efficiently forming these cells in the lab and then transplanting them could help type 1 diabetics better regulate their blood sugar — and genome-wide analysis of single cells among the mix has helped the scientists accurately distinguish and isolate beta cells from other classes of pancreatic cells.
“Cells don’t necessarily care about their history,” Klein says. “They may have to transition to one identity in one place and another identity in another.”