Living things must deal with a universe that is both regular and ever-changing: No day exactly mirrors the last, yet the sun and moon still appear at their appointed hours.
Cells contain their own seeming chaos, with countless molecules cooperating to produce subtle responses and behaviors. And in recent decades, a great deal of focus has specifically centered on the periodic patterns that underlie many cellular processes.
Oscillations — such as a pendulum’s swing or a ball’s bouncing on the end of a spring — are among the simplest and most common phenomena in physics, but researchers have come to appreciate their ubiquity in the biological world, too. Concentrations of molecules rise and fall, genes alternate between on and off, and circadian clocks keep time almost as well as human-made machinery. Together, these biochemical fluctuations are crucial for a blizzard of biological needs: timing daily activities, orchestrating cell division and movement, even mapping out parts of an embryo as it grows. Cells would be unable to function without them.
Synthetic biologist Michael Elowitz of Caltech discusses the importance of oscillations in cells, and how they are inspiring scientists to create cells with new functions.
PRODUCED BY HUNNIMEDIA FOR KNOWABLE MAGAZINE
Such patterns were harder to spot in years past because scientists analyzed whole populations of cells at a time and looked at averages, says synthetic and systems biologist Michael Elowitz of Caltech in Pasadena. But biochemists can now tag molecules in individual cells with fluorescent biomarkers and film their ebbs and flows. “More and more people started to look at individual cells over time and discovered that some of the most important systems in biology are not static — they’re really dynamic,” Elowitz says.
Some biochemical oscillations are simple: A few proteins or other organic chemicals go through a repeating pattern. Others are so complex that scientists have yet to map out their pathways. But their pervasiveness has drawn a great deal of attention from those seeking insight into biochemical behavior and researchers like Elowitz who hope to apply such knowledge by engineering novel functions into cells.
“All of these are self-organized,” says theoretical physicist Karsten Kruse of the University of Geneva in Switzerland, who coauthored an article about oscillations in the Annual Review of Condensed Matter Physics. “If you add the right components together, then they don’t have a choice — they must produce these oscillations.”
Here’s a look at some of the most well-studied and intriguing biochemical oscillations that emerge from the complexity of the cell to produce order.
Circadian rhythms in cyanobacteria
Daily activity cycles are important for survival in our 24-hour world. In 2017, the Nobel Prize in Physiology or Medicine went to researchers who unraveled the details underlying these rhythms in higher creatures. In contrast, single-celled organisms, such as light-harvesting blue-green algae or cyanobacteria, were once thought too simple and fast-dividing to harbor such clocks.
But keeping track of the sun is obviously important for organisms whose livelihood depends on light. Today researchers know that these life forms also have intrinsic circadian rhythms — and know a lot about how they function. Molecular geneticist Susan Golden of the University of California, San Diego, has helped to decode the molecular machinery regulating time in the cyanobacterium Synechococcus elongatus, and coauthored a description of the clock in the Annual Review of Genetics. The story goes like this:
The cyanobacterial circadian rhythm relies on an oscillation among three proteins: the enormous KaiC, which consists of two six-sided, doughnut-like rings stacked atop one another; its helper, the butterfly-shaped KaiA; and the component KaiB, which is usually inert but can spontaneously change to a rare, active form.
As the sun rises, wiggly molecular chains extending from the top of KaiC’s upper stack grab hold of little KaiA. Once bound, KaiA induces the immense KaiC to accept phosphate groups. Over the course of the day, more and more phosphate is added to KaiC’s top ring, stiffening it and causing its lower doughnut to deform.
By sunset, the lower ring has been so squished that it exposes a hidden binding site along its bottom. The rare active form of KaiB can now stick to this site, changing KaiC’s structure so it lets go of KaiA. As the night progresses, KaiC slowly gives up phosphates, eventually returning to its original state and releasing KaiB. The cycle takes about 24 hours.
And how does this oscillation cause rhythms in the cell’s biochemical activities? By cyclically activating a key gene-regulating protein named RpaA. RpaA switches on (or off) around 100 genes in S. elongatus. These genes, in turn, direct the cell’s metabolism and physiology — telling it, for instance, when it’s time to photosynthesize or burn sugar stores. Since RpaA activity peaks at dusk, the bevy of activities occur with daily cycles.
The cyanobacterium Synechococcus elongatus organizes the timing of activities such as photosynthesis with an internal clock. The clock oscillates between methodically adding molecules to a specific protein and then removing them.
PRODUCED BY HUNNIMEDIA FOR KNOWABLE MAGAZINE
Division in E. coli
Bacteria divide to reproduce, but an off-center partition will cause lopsided daughter cells, potentially leaving descendants understocked with the materials they need to survive. Not surprisingly, then, many microbes use molecular systems to split perfectly in half.
Perhaps the best understood is a team of three globule-shaped proteins called MinC, MinD and MinE that create waves of fluctuations in Escherichia coli.
The key component is MinC — in high concentrations, it blocks a protein that kicks off the process of division. But MinC doesn’t work solo. On its own, it will diffuse throughout an E. coli cell and stop division from happening anywhere at all. So MinC relies on MinD and MinE to tell it where to go.
MinD binds to the membrane at one end of the cell, painting the interior with clusters of itself. That attracts huge collections of MinC that come in and bind to MinD — blocking the molecular machinery that initiates division from setting up shop at that location.
Next comes the work of MinE. Lots of MinEs are attracted to the MinDs and they force MinD to undergo a small change. The result: MinDs and MinCs are kicked off the membrane. They move on to search for a place devoid of MinEs — like the other side of the bacterium — where they can bind once again to the cell membrane.
Then it happens all over: MinEs chase and kick off the MinD-MinC complexes again. Wherever MinD tries to stick to the wall, it gets booted out, and MinC along with it. The process generates a pulsation of Min proteins that moves back and forth between the cellular antipodes over the course of a minute.
Why does this cause the cell to divide right in the center? Because MinC spends the least time in the middle of the cell — giving the division machinery an opportunity to assemble there.
This wouldn’t be the case if E. coli’s sizing were different. By constructing synthetic rod-shaped compartments of different lengths and widths and introducing concentrations of MinD and MinE into them, biophysicist Petra Schwille of the Max Planck Institute of Biochemistry in Munich, Germany, and colleagues created beautiful videos of the molecules’ fluctuations. They showed that longer or shorter cells would allow the division site to be at other locations.
An oscillating wave of proteins within the bacterium E. coli helps it divide precisely in half.
PRODUCED BY HUNNIMEDIA FOR KNOWABLE MAGAZINE
Vertebrate segmentation
In the seventeenth century, Italian physiologist Marcello Malpighi used an early microscope to study developing chicken embryos and observe the formation of their spinal columns. More than 300 years later, modern researchers are still puzzling over the incredibly complex process that forms each vertebra and segment of the body. One key component: a clock-like oscillation that travels down the developing embryo.
“It’s easiest to think about it as an oscillator that gets displaced in space with a certain speed and direction,” says developmental biologist Olivier Pourquié of Harvard Medical School in Boston. Each time the embryo reaches a certain phase in the oscillation, it stamps out a segment. Then it goes through the cycle again, producing a second segment. And so on. “But because the oscillator moves, it will stamp the segment at a different position,” Pourquié says. “In this way, you can generate a sequential series of segments” along the length of a gradually extending body.
In embryos of vertebrates like fish, chickens, mice and humans, the future head is one of the first structures to appear. Later, bumpy segments called somites emerge, one by one, below the head, eventually giving rise to the spine, rib cage, skeletal muscles, cartilage and skin of the back. These ball-like pairs of somites are generated from tissue below the head when that tissue receives cues from two separate systems — called the wavefront and the clock — at the same time.
First, the wavefront. It involves two molecules, fibroblast growth factor (FGF) and Wnt, each of which forms a gradient, with their highest levels farthest from the head: a place near the tail that is constantly moving away as the embryo elongates. (An inhibitory substance called retinoic acid, produced by already formed somites, helps to keep FGF-Wnt activity toward the rear.) The two molecules set off a complex series of steps and act to inhibit somite formation. Somites appear right around the spots where they are least abundant.
Second, the clock component. That’s governed by a third molecule — called Notch — and the signaling pathway it sets off. Notch causes cells to oscillate between active, “permissive” states and inactive, “restrictive” states at a characteristic rate that varies from species to species. If the cells happen to be in a permissive state at a spot where the Wnt-FGF gradient has sufficiently weakened, a cascade of genetic activity tells cells in that region to gather into somites.
And as the body elongates and the tail moves farther from the head, the Wnt-FGF wavefront will move in a posterior direction, stamping out a line of somite segments with each tick of the Notch clock. (Read more about segment formation in this article in Knowable Magazine’s special report on Building Bodies.)
The developing mammalian embryo produces two somites, one each side of the future spinal canal, every time an internal clock “ticks.” The process is guided by a protein called FGF that is made by the tail end of the embryo and diffuses along its length, forming a gradient. Somite production occurs at a spot (the wave front) where the concentration of FGF is at just the right level when the clock makes a tick. The process repeats itself over and over, gradually building up segments, from which vertebrae and skeletal muscle are made. Two other molecules, Wnt and retinoic acid, also form gradients, and with FGF are key to telling tissues where they are along an embryo’s length.
Waving motion
Just like their multicellular kin, single-celled creatures need to move in order to hunt, escape predators or seek out light and nutrients. But getting around when you don’t have limbs can be a tough task. So cells that need to move, be they free-living or part of a multicelled creature, rely on various types of molecules to do the job. In certain cases, the action of these molecules can induce wave-like ripples on the cell’s surface, which the cell uses to skate forward.
Actin, a protein found broadly in nature, is key. The molecule, a major component of the mesh-like cytoskeleton, is involved in a slew of operations: mobility, contraction as cells divide, changes in cell shape and internal transport.
Along with colleagues, computational biologist Alex Mogilner of New York University in New York City has investigated how actin can drive waves that allow certain types of fish cells known as keratocytes to crawl around. Keratocytes are responsible for producing collagen and other connective proteins, moving to sites of injury and inflammation to assist in healing. They have often been used as model systems to study cell locomotion.
Normally, cells get around by protruding long, limb-like extensions and tottering forward like tiny, exotic aliens. But when they enter an especially sticky environment, their strategy changes and they no longer extend thin limbs, instead skimming forward using short ruffling motions of their cell membranes.
Beneath the membrane of a keratocyte, actin proteins are constantly assembling and disassembling into long filaments. In a highly adhesive environment, the cell membrane will sometimes stick to the external material, which tugs on the membrane as the cell tries to move. This tugging creates a small pocket right beneath the membrane that actin filaments can expand into.
An enzyme called vasodilator-stimulated phosphoprotein (VASP) will often be hanging around beneath the membrane, too. VASP binds to the actin and stimulates it to form even longer filaments and branches. If both VASP and actin are present in high enough concentrations, a cascade of actin filament-lengthening can begin. “When it starts, it’s like a fire starting,” says Mogilner.
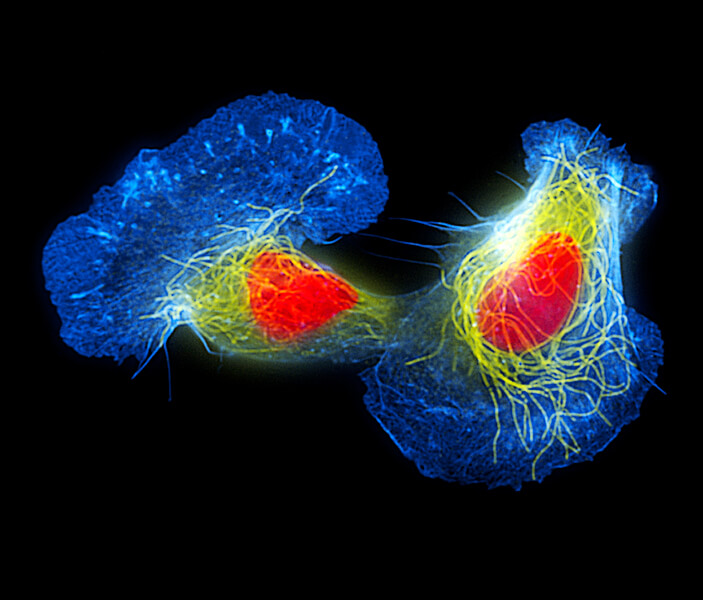
Fish skin cells called keratocytes are often used to study cell movement. That movement can involve waves generated by filaments of actin, a component of the cell’s cytoskeleton. In this colorized image, actin is shown in blue and microtubules, also part of the cytoskeleton, are shown in yellow. (The nucleus is colored orange.)
CREDIT: DR TORSTEN WITTMANN / SCIENCE SOURCE
The elongating filaments push on the tight cell membrane, producing a bump that gives the actin chains room to grow even more, and bind more VASP. Tension in the membrane causes it to sway like an audience doing “the wave,” sending the cell skating in the wave’s direction. The actin filaments beneath the membrane grow sideways as well as forward, helping to push the wave along. At the original spot where the wave began, the actin filaments will have used up all the available VASP, preventing further lengthening. The sticky external environment adhering to the taut membrane also dampens the wave at the origin spot.
“In a way, VASP proteins are like trees, actin filaments are like fire, and adhesions and membrane are like water: At the back of the wave, trees are all burnt and drenched in water, and the fire stops,” says Mogilner. But at parts of the membrane far from the wave’s origin, high concentrations of actin and free VASP will still exist, often leading to a new wave that begins where the previous one was extinguished.
It’s still unclear just how keratocytes choose what direction to move in. Presumably, says Mogilner, the leading edge of a cell is oriented toward some external cue, like a chemical gradient from some food. Also poorly understood are the benefits of this particular mobility tactic. “In some cases, it’s not obvious why waves are better than other mechanisms,” says Kruse, whose work on cytoskeleton dynamics focuses on theoretical descriptions of cell movement and division.
Some researchers have suggested that the wave-like motion might help cells get around small obstacles that they would otherwise run into head-on. Or maybe it’s prudent for them not to overextend their limb-like protrusions in certain environments.
A synthetic cellular circuit
When Caltech’s Elowitz was in graduate school at Princeton University in the 1990s, he often got frustrated by diagrams showing the inferred interactions of genes and proteins, with their many unknowns and arrows going every which way. “I just became convinced that if we really want to understand these things we need to be able to build them ourselves,” he says.
Along with his advisor, Stanislas Leibler, he created a synthetic genetic oscillator in order to show that a simple biological system could be programmed and built from scratch. Called the repressilator, it consists of a tiny loop of DNA with three genes on it. They carry instructions for making three proteins called repressors, each of which binds to the next gene and turns it off.
And here’s where it got fun. In their construction, the first gene produced a repressor protein, LacI, which would shut off the second gene, called tetR, whose product would shut off the third gene, cI, whose product would shut off the first gene.
“It’s like a game of rock, scissors, paper,” says Elowitz. “The first repressor turns off the second one, the second turns off the third one, and the third turns off the first one.” Once the first gene is turned off, the second gene can turn on, and thus turn off the third gene. And then the first gene can turn on again — and on and on.
To watch the circuit run, Elowitz included a fourth gene that would cause E. coli to light up bright green — but only when it was turned on by one of the three repressors. Placed inside E. coli, the repressilator causes the microbe and its descendants to flash green fluorescent light with a period of around 150 minutes.
Scientists created a tiny protein-making circuit and stuck it in bacteria. As the production of the three proteins rise and fall, the bacteria rhythmically pulse with green light.
PRODUCED BY HUNNIMEDIA FOR KNOWABLE MAGAZINE
Beyond simply showing that such circuits could be created, the research provided insight into the noise of biological systems. E. coli did not turn out to be a perfect little deterministic machine, says Elowitz. When loaded with the repressilator, some daughter cells flashed more strongly or weakly than others, suggesting that there is a great deal of variability inherent in their biochemical workings.
Studies have continued on the system and, in 2016, a team at Harvard University and the University of Cambridge significantly improved the precision of the circuit, allowing much larger numbers of daughter cells to flash in sync.
The field of synthetic biology has grown rapidly in the two decades since Elowitz’s early work, and now offers a plethora of interesting applications, including novel proteins and enzymes for medicine, biological sensors and even cells that perform calculations like living computers. Being able to fine-tune biochemical oscillations — with far more exquisite precision than can be found in natural systems — will be crucial to building future synthetic biological products, says Elowitz.
“Out of physics, we have electronics and electrical engineering,” he says. “We’re just beginning to learn these principles of genetic circuit design, and I think we’re at an interesting moment.”
Video Transcript:
Michael Elowitz (synthetic biologist, Caltech): “The cell, the living cell, is really the building block of all life. And I like to think of it as kind of the most amazing device we know about.
“The cell is a little tiny object, you know, maybe a few microns, 10 microns 15 microns, in size; very small — squishy, we usually think it’s squishy — but this little device can do amazing things. It can allow itself to divide and make copies of itself so it can proliferate. Those individual cells can talk to each other in incredibly precise ways. They can remember things; there’s memory in cells, and then they can develop into multicellular organisms like ourselves.
“So what we’re going to talk about today is one of my favorite phenomena in all of biology, which is oscillations.
“What is an oscillation? You have some pattern of behavior that’s repeating over and over again. When you go to the level of the single cell, look at that individual cell and you look at the, let’s say, the concentration of a particular protein that does something in the cell. You find a lot of times that it goes up and it goes down, so the proteins are made, stay there for a while and go away, made and then go away, over and over again. That’s an oscillation — a rhythmic change in the level of the protein.
“Some of these oscillations do really fundamental things. The cell cycle is an oscillator. That’s a sequence of events that every cell goes through as it replicates its DNA and then divides that DNA into two daughter cells and then does it again.
“This video is trying to explain what’s going on when a bacterial cell divides. This is the most important thing. If the bacterial cell can’t divide, they wouldn’t be so successful. So they’ve got to kind of do this all the time over and over again. Their challenge as a cell is to divide in the middle. If they miss the middle, they run the risk of not having one copy of each chromosome of their DNA go to each daughter cell. So they have this challenge, which is finding their own middle point and then squeezing off right in the middle. And what we’ve learned is that there’s a molecular system of a few proteins that interact with each other, swish back and forth from one end of the cell to the other, and use that to actually help the cell find that midpoint.
“I think it’s a beautiful case in which you have an unexpected discovery that somebody made really just by looking at these proteins in the microscope directly for the first time. So I think this system really kind of epitomizes a lot of the trends and themes and forces that are happening right now in biology.
“There’s also a circadian clock. That means that every cell in your body actually has a clock circuit that oscillates with a 24-hour rhythm, and that those 24-hour clocks are actually guiding your body through its normal circadian cycle during the day. So that means that even if we were to put you in a cave with no signals, no temperature changes, no changes in light, you would still have this 24-hour behavioral rhythm.
“One of the things that was really astonishing, when biologists discovered it, was that actually even a single-cell organism can also have a circadian clock. So that’s true for cyanobacteria. These are photosynthetic bacteria, and they need this clock because they alternate between phases when they photosynthesize and phases in which they fix nitrogen, and those two activities are incompatible — they can’t do them at the same time in the same cell, and so that’s really why they have a circadian clock, or one of the main reasons.
“The key point about the cyanobacteria clock in particular: It’s so amazingly simple. I don’t think anybody would have anticipated that you could build such a precise clock out of just three proteins, and that that clock could operate outside of the cell altogether, can be reconstituted in a test tube. That was just amazing.
“So this example here in the cyanobacterial case really is one in which you can actually understand how a sequence of molecular events, just little interactions between molecules, actually gives rise to a change in behavior at the level of a whole cell over time, in a circadian way. So it’s a great example of, really, what you can do in biology, which is connect molecules interacting with each other to the behavior of a cell as a whole.
“When I was in grad school, I just wanted to actually build our own clock out of genes and proteins, put it into a cell, and really show that it was sufficient to give you oscillations, and then to really understand what kinds of designs are sufficient for oscillations, to really understand that directly.
“So what you’re seeing in this animation: The circle is literally a circular piece of DNA. So that circular piece of DNA contains the three genes of the repressilator. The design itself really is a kind of rock-scissors-paper circuit. That really is exactly what it is. The first gene, which we can call “rock,” turns off the second gene, which we’ll call “scissors,” that really turns off the third one that really turns off the first one. So the difference is, unlike when you might play rock-scissors-paper with your friend in order to settle some kind of decision about who’s going to do what, here the rock, scissors and paper are kind of separate entities that are each influencing each other. And so that’s the beauty of this circuit: It can’t do anything else except oscillate.
“What was most surprising is that it worked, because while I was going through that process, I told a lot people what I was trying to do and there was a really polarized response. So I think a lot of people, even very experienced biologists, felt that it just wouldn’t work. Not for any specific reason, but just because the cell is complicated. And some simple design that you dream up and should write down a piece of paper may not work in the complicated environment of the cell, where there’s tons of different molecular interactions at play.
“The key idea is the idea that you’re building new behaviors in cells — from scratch. Why try to program new cellular behaviors? If you just want to understand how cells not only do work, but how they could work. What’s possible with these amazing genes, proteins and other biomolecules. It really changes our notion of what biology is. It’s not just a collection of organisms, it’s really a system for creating new functions in cells. So I think there’s a whole exciting world coming, I think, in the future, in which we’ll be able to think of a circuit as a therapeutic and start thinking of it as something that programs therapeutic activities in cells and kind of unlocks all those capabilities.
“It’s really like thinking about biology as a set of Legos that we have and thinking about not just, you know, if you think about with Lego kits you can sometimes build a specific thing, but you could use those Legos to build many things. And that’s what biology is about. I think there’s just a fundamental curiosity to understand what’s possible.”