For decades, Piri Welcsh has had professional and personal stakes in understanding the genetics of breast cancer.
In the 1990s, the molecular geneticist participated in an international race to clone BRCA1, the first gene linked to breast cancer risk, and she works to this day in the lab of pioneering breast cancer geneticist Mary-Claire King at the University of Washington.
And then there’s Welcsh’s own family. Her grandmother died of breast cancer, her mother is a breast cancer survivor, and her aunt was just diagnosed with breast cancer.
Since the days of the BRCA1 discovery, researchers have identified more than 100 genes that, in certain forms, raise breast cancer risk. And yet that still leaves many people with elevated familial risk seeking answers. “Some families, like mine, are unsolved,” says Welcsh, who is vice president of education for the support group FORCE: Facing Our Risk of Cancer Empowered. “They seem to have something genetic being passed through generations, yet no mutation identified.”
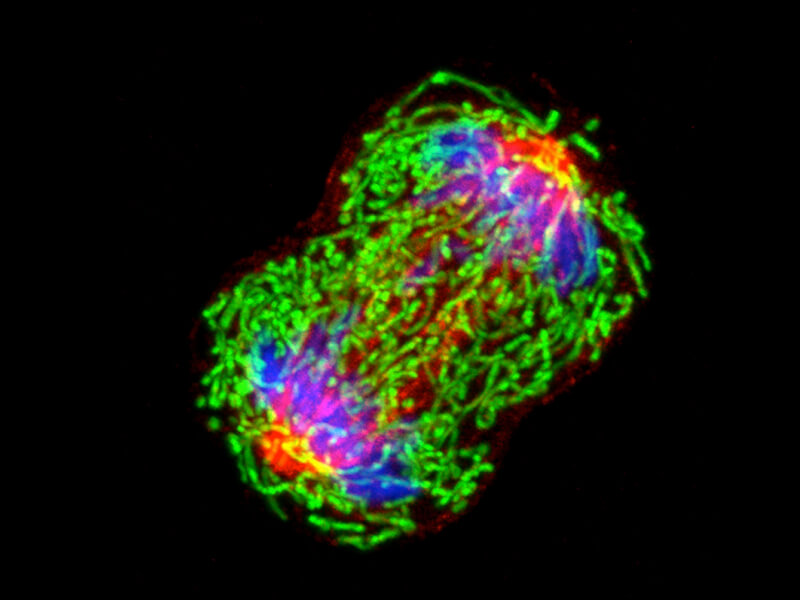
Light-microscope image of a dividing breast cancer cell, showing chromosomes (blue), the mesh of tubules that pull the chromosomes to the poles (red) and mitochondria (green).
CREDIT: NATIONAL CANCER INSTITUTE / UNIV. OF PITTSBURGH CANCER INSTITUTE
Researchers hope to offer explanations to many women like Welcsh by broadening their knowledge of the genes and gene variants involved in breast cancer risk through a focus on tiny DNA differences between one person’s genome and the next — variants known as SNPs, or single nucleotide polymorphisms. By so doing, they aim to come to a better understanding of the causes of breast cancer and develop more comprehensive screening panels — and, down the road, perhaps new and more bespoke treatments.
Familial mysteries
There are more than 3 million women in the US with a history of breast cancer, and an additional 266,120 cases are expected to be diagnosed this year. Non-genetic risk factors such as reproductive and menstrual history are well established and account for the lion’s share of breast cancer cases, which are mostly sporadic. But about 5 to 10 percent of breast cancers are genetic. Most of them are linked to variants of the breast cancer genes BRCA1 and BRCA2, while another few percent are linked to variants in other genes.
An additional 15 to 20 percent of women with breast cancer have a family history of the disease — with two or more first- or second-degree relatives who have been diagnosed with breast cancer — but no genetic cause yet identified.
This pie chart illustrates the proportion of breast cancer cases that run in families, compared with sporadic cases. Cases rooted in genetics are divided into two categories based on the strength and pattern of inheritance. The “hereditary” class is reserved for cases of breast cancer that are inherited with an autosomal dominant pattern — meaning that the trait passes along as if it were a dominant gene located on the non-sex chromosomes. The well-known BRCA1 and BRCA2 gene variants are examples.
To try to understand where the heightened risk is coming from in this so-called “familial” category, in recent years researchers have turned to GWAS (genome-wide association studies) — technology that scours people’s genomes to find SNP variations that are more frequent in people with breast cancer than those without it. This, in turn, can identify genes involved in risk.
So far, more than 150 SNPs have been associated with breast cancer. The vast majority of these appear to increase risk by just a fraction of a percent (compared with up to 70 percent for certain BRCA variants, and on the order of 10 percent for other known variants in other genes). But if many such SNPs were present in the same person’s genome, their effect could add up to significant risk, researchers say.
Most of these minor variants lie not within the core of a gene itself but in regulatory areas that determine how and when a gene is turned on and off, according to a 2017 report by an international group of researchers called the OncoArray Consortium. The group surveyed DNA data from more than 100,000 women of European and East Asian ancestry who had breast cancer and compared them with a similar number of cancer-free individuals, identifying 65 new genome regions associated with breast cancer risk.
There are likely many more to be identified, says study coauthor Montserrat García-Closas, an epidemiologist at the National Cancer Institute. GWAS studies can pick up only the highest signals — variants contributing the greatest effects — but don’t detect ones below a certain threshold, she says. More precisely, the OncoArray authors calculated that the variants they could detect explain just 18 percent of the elevated breast cancer risk seen in women in the familial class, while ones below the threshold could explain an additional 23 percent.
A growing number of gene variants have been linked to an elevated risk of breast cancer. Risk can be raised by different amounts depending on the specific variant of a given gene that a person carries. To understand the many inherited cases that remain unexplained, scientists are turning to tiny, single-letter differences known as SNPs. Though each SNP may raise risk only slightly, a combination of several may as much as double risk if they affect the same, or related, biochemical pathways.
Identifying these new variants is important for several reasons, oncologists say. First of all, it provides patients and their doctors the approximate risk of a woman developing breast (and often other) cancers. In some cases, it gives physicians information about the biology of a patient’s cancer and can be used to devise screening protocols, prevention tactics and treatment methods.
Recently, commercial labs such as Myriad Genetics and Ambry Genetics began offering to test for certain SNP variants when running the traditional panel of known breast cancer genes. This may be jumping the gun, says García-Closas — because studies calibrating and validating how much these variants elevate risks are still underway and it could be three to five years before they’re strongly validated. “I would take, with a little grain of salt, the value of the current risk scores available,” she says. “I think this is still in the early stages and I think they will get better.”
Before physicians decide to use variants in panels, they generally want to know that those variants are adequately understood, raise disease risk by a significant enough amount and make a tangible difference to clinical management, adds Katherine Nathanson, a clinical geneticist at the University of Pennsylvania’s Perelman School of Medicine.
“BRCA1 and BRCA2, for example — we’ve known about them for a long time, we really know what kind of risks they are associated with, we know how to interpret those mutations,” she says. “But there are genes that have been more recently described, or for which we don’t fully understand what the cancer risks are, or the risks are lower, but are also sort of grouped together on those panels.”
Once such facts are solidified, including them in a panel may be an asset to at-risk women seeking health insurance coverage for additional breast cancer screening. The National Comprehensive Cancer Network has guidelines for managing women who carry genetic variants identified through panels, including more frequent breast exams and more sensitive screening methods such as breast MRIs. Individuals at especially high risk may be offered prophylactic interventions such as mastectomy or chemoprevention drugs like tamoxifen.
But the identified risk has to be high enough and established before insurance companies will pay for such treatments, says Rachel Shapira, a genetic counselor at UCLA Health in Los Angeles. Companies generally require a lifetime risk score — derived from genetic tests or family history assessments such as the Tyrer-Cuzick risk model — of over 20 percent before covering annual breast MRIs in addition to annual mammograms. And many SNPs are too new for there to be clinical or insurance guidelines that address them.
Typical of the questions getting bounced around, Shapira says, is “if I have a patient — their Tyrer-Cuzick score is 16 percent, but their risk score with SNPs is 30 percent — will their insurance pay for anything? And we don’t know yet.”
There are millions of places in the human genome where a DNA letter can vary from one person to the next. These differences are known as SNPs (single nucleotide polymorphisms). Researchers are looking for SNPs linked to an elevated risk of breast cancer in order to help explain cases where breast cancer runs in families but no genetic cause is yet known. In this example, Person 1 has a SNP that has been linked to elevated breast cancer risk in population studies; the other two individuals have SNPs that are not.
Scientists are continuing to study identified breast-cancer-linked SNPs and find new ones with smaller effects. Since some SNPs might not, individually, raise breast cancer risk by much, homing in on risk-raising combinations could be key, says Chad Myers, a computational biologist at the University of Minnesota. Such studies aren’t easy because they require very large groups of women, he says. “For each person, you have 500,000, sometimes one million, different SNPs you measure. That’s like a half-trillion combinations to look at.”
Myers and his team tried a more systematic approach. They chose to study sets of genes that are known to work together in the same or related biochemical pathways. The idea is this: Say that disrupting a certain pathway raises breast cancer risk. In that case, individual SNPs in genes in that pathway might compromise it only slightly. But when several different SNPs affecting the pathway at different spots occur together in the same genome, they could damage it significantly. And because the human body has lots of redundancies — a physiological need can often be met by more than one biochemical pathway — several SNPs that impair two related pathways might similarly carry a lot of clout.
In a 2017 study of nearly 8,000 women with breast cancer and more than 8,500 without, Myers and his colleagues did, indeed, find evidence that combinations of SNPs within one pathway, and especially two related pathways, can raise the breast cancer risk significantly above what a single SNP could do — roughly doubling lifetime risk in some cases, Myers estimates. The study pinpointed 25 biochemical pathways involved in breast cancer risk, 16 of which had never before been implicated. These could therefore provide new insights into the mechanisms behind increased risk.
Researchers are just in the early stages of understanding what all these SNPs do to raise breast cancer risk. Will they ultimately offer answers to people like Welcsh, who still hopes to find her genetic link so the family can plan preventive actions? Welcsh wonders about that. “It could be that just in my family, there is a mutation in a gene, [and] that no one else in the world has a mutation in that gene – it could be unique to one family,” she says.
But others are more hopeful that SNPs stand to open new doors to understanding. “We’re just scratching the surface,” Nathanson says.